Impact of antibiotics on the gut microbiota, does it matter?
Overview
By Pr. Francisco Guarner
Digestive System Research Unit, University Hospital Vall d’Hebron, Barcelona, Spain
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections

About this article
Author
Massive misuse of antibiotics may lead not only to antimicrobial resistant infections but also to the spread of non-communicable, chronic diseases. The use of antibiotics and vaccines have done more to extend life expectancy than any other medical innovation. Antibiotics are among the most recognized medical milestones according to experts appointed by the British Medical Journal. [1] After the introduction of measures for the prevention and treatment of infectious diseases, changes were gigantic. Infections typically caused 30% of all deaths, mostly in children aged under 5 years, but at the end of the 20th century less than 4% of deaths were due to infection.[1] However, two major concerns have recently emerged. First, treatments for a growing number of infections become less effective due to resistance. Antimicrobial resistance is now a major threat to human health, and its link with overuse of antibiotics is well documented.[2] Second, antibiotics meant to kill pathogenic microbes have had unintended consequences for the human microbial ecosystem, including changes that may be difficult to reverse.[3] The human body is home to a complex array of microbes known as the microbiome or microbiota, which play an important role in health. Microbiota alteration and the accompanying loss of functional attributes might result in the microbial communities of people living in industrialized societies being suboptimal for health.[3] These issues require awareness by the medical community and straightforward guidelines from health policy makers.
Impact of massive use of antibiotics
According to the WHO 2018 report,[2] the overall amount of antibiotics consumed by humans is well above 6,500 tonnes per year (data from 65 countries; China and US not included). A median of 18 out of 1,000 inhabitants consume every day a defined dose of antibiotics, which means that 139 million doses are consumed every single day of the year. The median is lower in African countries (12 out of 1.000) than in Europe (17,8) or America (18,2), while infections cause up to 36,6% of total deaths in Africa but only 2,7% or 4,5% in Europe or America. Low-income countries still have high mortality rates from infectious diseases but low rates of antibiotic use. Limited access, use of wrong drugs or wrong treatment schedules, can contribute to resistant infections arising in low-income countries, like tuberculosis.
In developed countries, as many as half of all antibiotic prescriptions can be considered inappropriate.[2] Unnecessary antibiotic consumption accelerates the development of resistances, and multi-drug resistant strains of Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, etc., are on the rise.[2]
Hailed as one of the greatest medical advances of the 20th century, antibiotics have saved millions of lives. But they also have an impact on our microbiota by inducing a dysbiosis. Let’s take a look at this ambivalence role:
The ambivalent role of antibiotics
Antibiotic-induced dysbiosis
Although most courses of antibiotics result in no immediate, obvious side-effects, there is concern about collateral damage altering the composition of the gut microbiota and its functions [3]. Antibiotic-associated diarrhoea is the most commonly recognized complication of antibiotics, and develops in 15 to 25% of patients. Most episodes of antibiotics-induced diarrhoea are mild and self-limited. However, an increasing number of cases develop more severe forms, including Clostridioides difficile associated diarrhoea. Antibiotic-induced disturbances promote C. difficile spore germination within the intestine, overgrowth of vegetative forms and toxin production, leading to epithelial damage and colitis. Clinical presentation ranges from self-limiting diarrhoea to toxic megacolon, fulminant colitis and death.[4]
From birth onwards, the human gut microbiota rapidly increases in diversity during the first 3 years of age, before stabilizing to an adult-like state. Thereafter, the core composition is stable but bacterial abundances may fluctuate in response to external factors (diet, drugs, travel, etc.). Studies have shown that the effects of antibiotics result in very large shifts in relative abundances. In patients on β-lactams or quinolones, the core microbiota fell from 29 to 12 taxa, the total number of observed taxa decreased by 25%, and there was a shift from Faecalibacterium to Bacteroides as the dominant genus.[5]
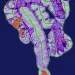
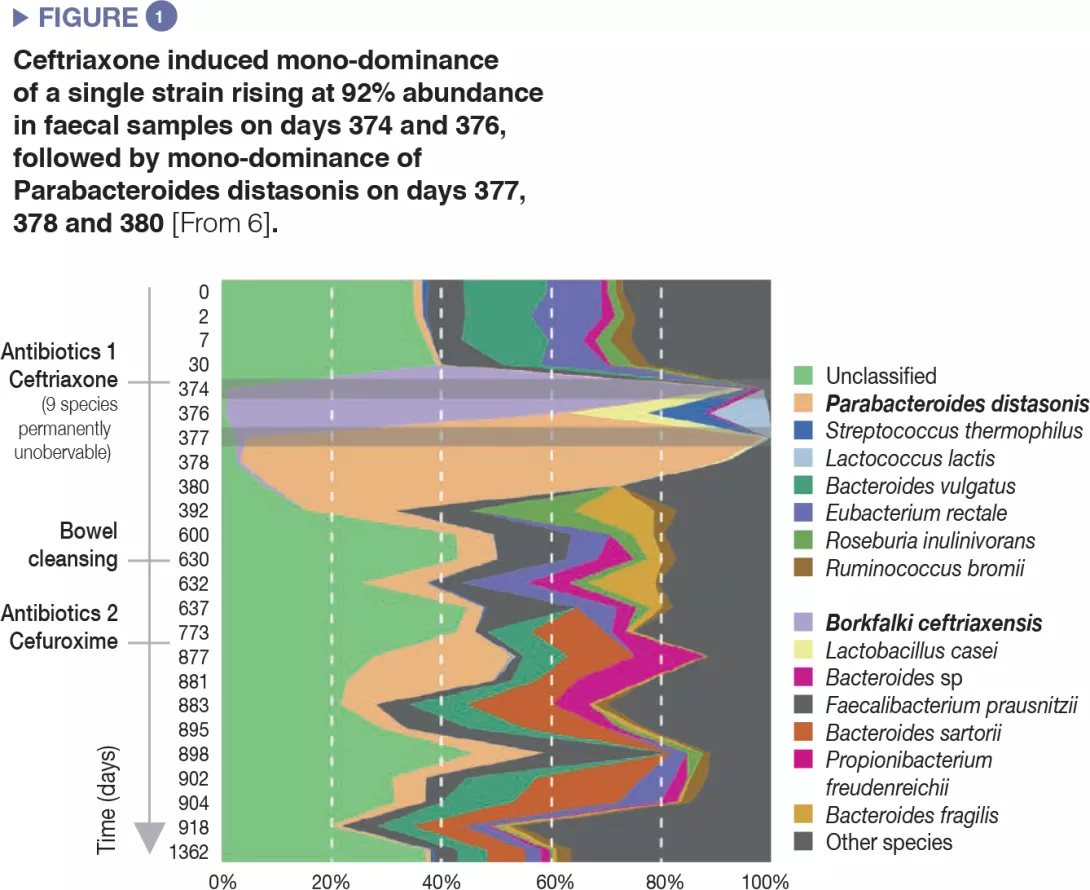
The use of antibiotics induced a decrease in microbial diversity (loss of richness in the ecosystem) and overgrowth of resistant species, which resulted in overall increase of microbial load, i.e. number of bacteria per gram of faeces.[5] Extreme cases of antibiotic-induced overgrowth have been reported showing mono-dominance of a single strain that bloomed to 92% relative abundance in faecal samples after intravenous ceftriaxone treatment.[6] (Figure 1).
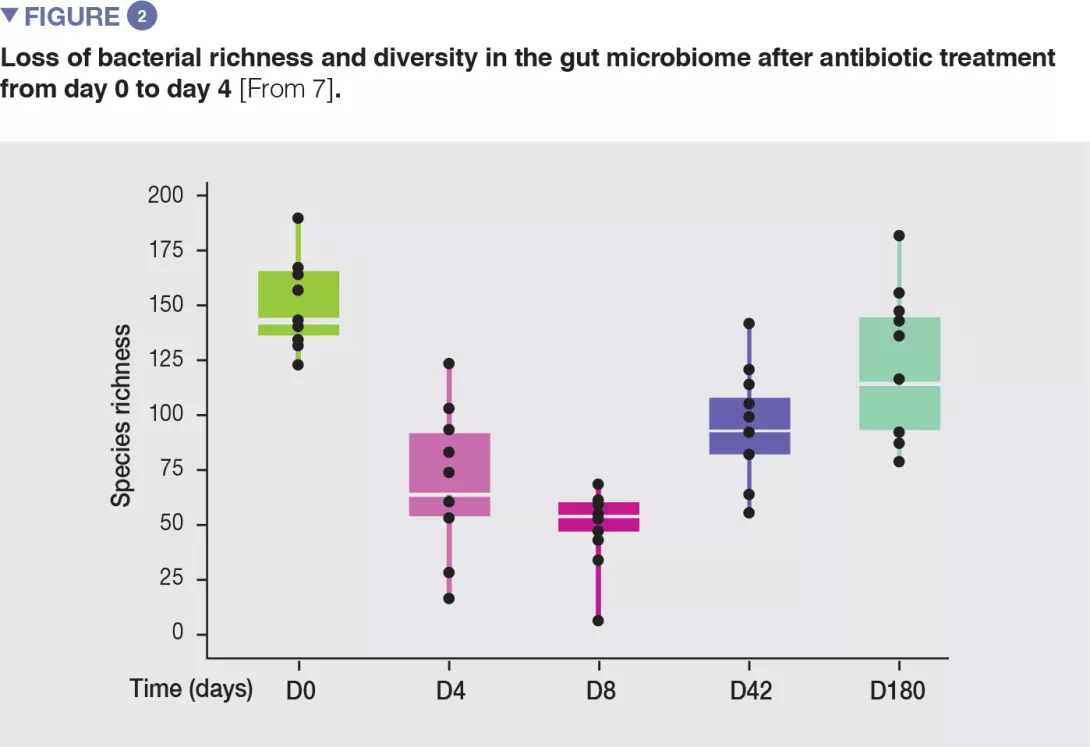
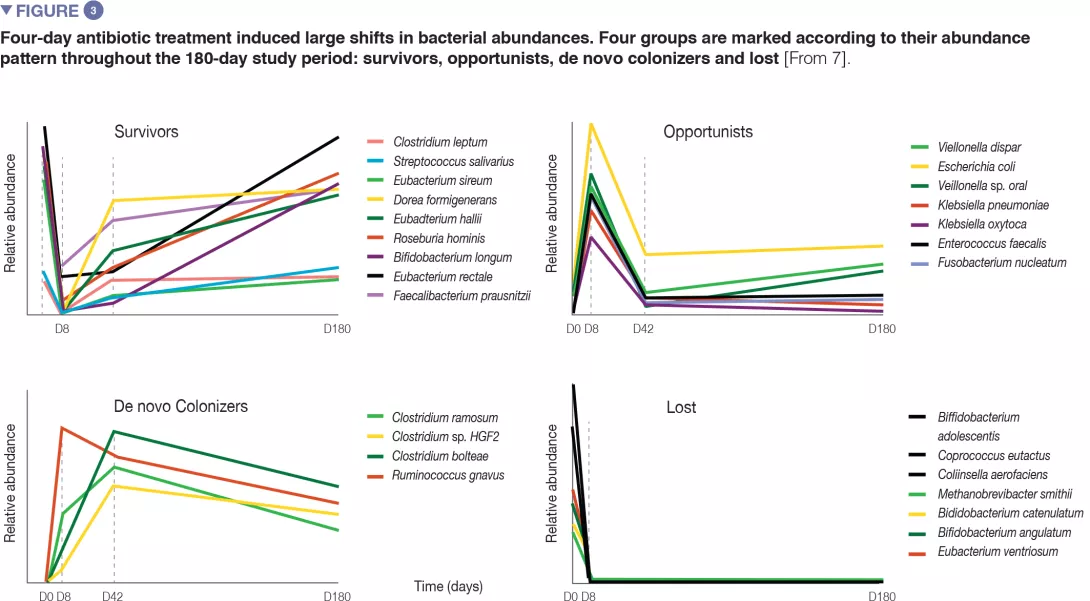
In healthy volunteers, a 4-day antibiotic intervention led to blooms of enterobacteria and other pathobionts (Enterococcus faecalis, Fusobacterium nucleatum), and to the depletion of Bifidobacterium species and butyrate producers.[7] The gut microbiota only recovered to near-baseline composition within 1.5 months, although 9 common species, which were present in all subjects before the treatment, remained undetectable after 180 days. (Figures 2 and 3).
Dysbiosis is a compositional and functional alteration in the microbiota that perturbs the microbial ecosystem to an extent that exceeds its resistance and resilience capabilities.[8] The functional impact of antibiotics on short chain fatty acid producers, butyrate in particular, may have long-term consequences because of the rupture of the symbiotic balance between microbiota and host. Failure to produce butyrate increases the flow of oxygen towards the mucosa and perturbs the micro-ecosystem in a way that favours the survival of oxygen-resistant bacteria (enterobacteria) and precludes recovery of butyrate producers like Faecalibacterium, which are strict anaerobes.[9] Such changes critically affect the resilience capacity of the ecosystem and perpetuate the imbalance towards chronicity.
Unintended side-effects of antibiotics on the gut microbiota and the accompanying loss of functional attributes might result in rupture of the symbiotic balance between microbiota and host.
The resistome
The resistome is the collection of all bacterial genes that directly or indirectly contribute to antibiotic resistance. Resistance genes do not seem to have been selected in response to recent exposure to antibiotics. Antibiotics date back hundreds of millions of years, so is resistance, and the number of genes in the resistome is a reflection of the continuous co-evolution of antibiotic-producing and target organisms. Composition of the resistome and prevalence of resistance genes in human-associated bacteria adapt to selective forces derived from human action.
Species that harbour β-lactam resistance genes are positively selected during and after antibiotic consumption.[7] Likewise, harbouring amino-glycoside resistance genes also increases odds of de novo colonization. Antibiotic resistance gene carriage modulates the recovery process after antibiotic consumption.[7]
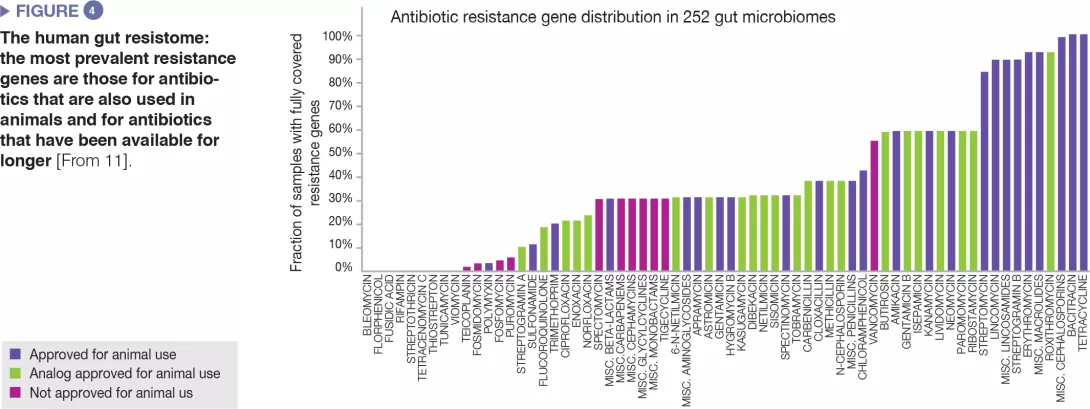
The human gut microbiome harbours a diverse repertoire of antibiotic resistance genes, which can be investigated by molecular sequencing technologies.[10] A study on 252 human faecal samples from different countries found that the most prevalent resistance genes in the microbiome are those corresponding to antibiotics also used in animals and to antibiotics available since a long time (Figure 4).[11] Country-level data on antibiotic use in both humans and animals matched the observed country-specific differences in prevalence of resistance genes. Altogether, the data suggest a positive correlation between exposure to antibiotics and prevalence of antibiotic resistance genes.
Some antibiotic resistance genes are readily exchanged between bacteria through horizontal gene transfer. Studies have shown that under antibiotic-induced stress, rising opportunist bacteria spread resistance genes among the microbial community. A longitudinal study of the gut microbiome in Finnish children observed that the use of antibiotics promoted the expansion of antibiotic resistance genes in the gut, due to overgrowth of bacteria harbouring resistance genes and increased mobilization of resistance genes by plasmids.[12] Antibiotic resistance genes carried on microbial chromosomes showed a peak in abundance after antibiotic treatment followed by a sharp decline, whereas abundance of resistance genes carried on mobile elements persisted long after antibiotic therapy ended. This might be explained by the fact that episomal genes can be broadly distributed across multiple species by horizontal gene transfer.
Under antibiotic-induced stress, opportunist bacteria spread resistance genes among the gut microbial community. The human gut microbiota is an accumulator of resistance genes potentially providing them to pathogens.
The human gut microbiota may be the most accessible reservoir of resistance genes to pathogens. Early life antibiotic treatment is associated with reduced microbial diversity but also with an increased risk of antibiotic resistance development.
Probiotics may prevent the overgrowth of resistant species during antibiotic treatment and minimise the spread of antibiotic resistance genes.
Antibiotics and risk of disease
Perturbations of the gut microbial ecosystem during early life combined with genetic susceptibility may have a long-lasting impact on the immune system leading to disease or predisposition to disease later in life. Indeed, it has been shown that inflammatory bowel diseases, metabolic disorders (type 2 diabetes, obesity), and atopic diseases are associated with an altered composition of the gut microbiota.
A leading hypothesis regarding the pathogenesis of inflammatory disorders is that alterations of the gut microbiota caused by repeated exposure to antibiotics trigger inflammation. Infants receiving antibiotics before one year of age were found to have a 5.5 times higher risk of developing IBD than unexposed children.[13] Likewise, antibiotic exposure in the first 2 years of life, when host adipocyte populations are developing, are associated with a diagnosis of childhood obesity.[14] Reduced gut microbial richness has been associated with increased adiposity, insulin and leptin resistance, and a more pronounced inflammatory phenotype.
Conclusion
Despite the resilience of the intestinal microbiota, the spread of antibiotic resistance genes is now a major threat to human health, and overuse of antibiotics seems to be the leading cause. In addition, there is growing evidence linking fragility of the human microbiota in industrialized countries with the coincident spread of non-communicable, chronic diseases [3]. Antibiotic exposure is again an obvious cause for such microbiota derangement. Shifting current trends towards more sustainable medical practices is a major challenge for public health in the 21st century. Unquestionably, restricted and rational use of antibiotics is the best and most efficient way of preventing detrimental imbalances of the human gut microbiome. Interestingly, use of a probiotic with proven efficacy for prevention of antibiotic-induced diarrhoea has been shown to constrain the overgrowth of resistant species during antibiotic treatment.[15] Potentially, this strategy could also minimise the spread of antibiotic resistance genes.
Each year, since 2015, the WHO organizes the World AMR Awareness Week (WAAW), which aims to increase awareness of global antimicrobial resistance.
Held on 18-24 November, this campaign encourages the general public, healthcare professionals and decision-makers to use antimicrobials carefully, to prevent the further emergence of antimicrobial resistance.