Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease
Commented articles - Adult's section
By Pr. Harry Sokol
Gastroenterology and Nutrition Department, Saint-Antoine Hospital, Paris, France
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections
Bacteriophages infecting bacteria, illustration. A bacteriophage, or phage, is a virus that infects bacteria.
About this article
Author
Commentary on the original publication by Clooney et al. (Cell Host & Microbe 2019) [1]
The human gut virome is thought to significantly impact the microbiome and human health. However, most analyses have been performed on a limited fraction of known viruses. Using whole-virome analysis on a published inflammatory bowel disease (IBD) cohort and an in-house ulcerative colitis dataset, the authors shed light on the composition of the human gut virome in IBD beyond this identifiable minority. They observed IBD-specific changes to the virome and increased numbers of temperate phage sequences in individuals with Crohn’s disease. Unlike prior database-dependent methods, no changes in viral richness were observed. Among IBD subjects, the changes in virome composition reflected alterations in bacterial composition. Furthermore, incorporating both bacteriome and virome composition offered greater classification power between health and disease. This approach to analyzing whole virome across cohorts highlights significant IBD signals, which may be crucial for developing future biomarkers and therapeutics.
What do we already know about this subject?
The virome is probably one of the main forces that shape the human gut microbiome, but it may also be its least understood component. The virome is composed mostly of bacteriophages (phages), which play a key role in many microbial ecosystems by stimulating diversity, helping to replenish nutrients and facilitating horizontal gene transfer. Understanding the role of bacteriophages in the structures of microbial communities will be essential if we are to understand and control the alterations in the human gut microbiome that are associated with various diseases. Many gut bacteria (and potential phage hosts) are difficult to grow in the laboratory, which means that virome analysis is highly dependent on metagenomic sequencing and bioinformatic approaches.
Key points
-
A large majority of the gut virome cannot be studied because it is absent from the databases.
-
The database-independent method described here allows analysis of the whole gut virome.
-
In IBD, the gut virome is altered, less stable, and dominated by temperate phages.
-
The gut virome could be used as biomarker or therapeutics in the future.
However, phages lack universal marker genes (like the 16S rRNA gene found in bacteria), and there is a lack of taxonomic information with sparse databases, which means that methods are needed that are independent of databases. The first metagenomic studies revealed the diversity of the human gut virome, but could only classify a very small fraction (2%) of the DNA sequenced.[2] Improvements in high throughput sequencing technologies now make it possible to analyze the virome with an unprecedented level of detail. It has been confirmed that the virome is incredibly diverse, that the majority do not align to any reference virus sequences in databases (referred to as viral dark matter), and that the composition is unique to individuals.
Although the etiology of IBD remains unclear, these diseases are multifactorial and associated with alterations in the gut microbiome. Emerging data now indicate that the gut virome is altered in IBD [3] with greater overall diversity and an increased relative abundance of the order Caudovirales. However, almost all the results have been based on changes in the composition of the identifiable fraction of the virome, which can represent as little as 15% of the dataset.[3] This limits our overall understanding of the virome and hinders the identification of potential disease biomarkers. An analytical method independent of databases is essential if we want to fully characterize the alterations in the gut virome.
What are the main insights from this study?
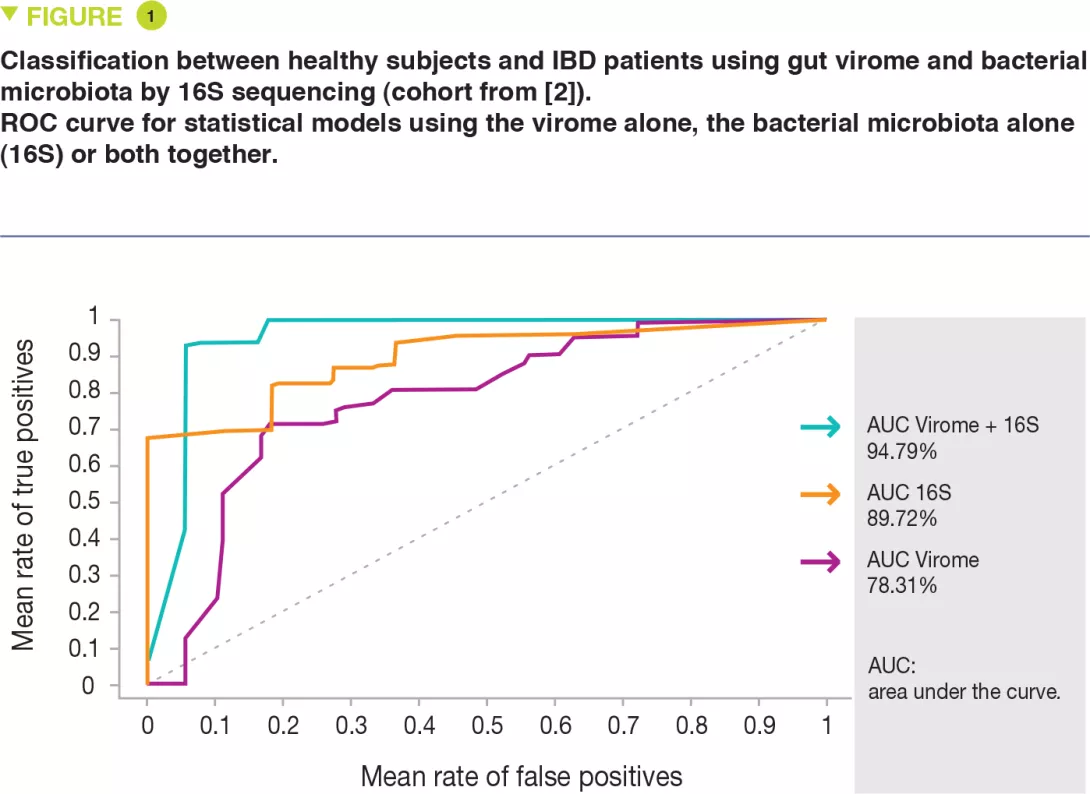
The authors of this study re-analyzed a set of published keystone data [3] from a Crohn’s disease and ulcerative colitis cohort and from healthy controls. The problem of high interindividual variation was overcome by using protein homology and a specific algorithm (Markov cluster) to group the viral sequences into presumed taxonomic ranks. This made it possible to describe changes in composition across the whole virome beyond the identifiable minority. The authors suggest that unlike the core virome of healthy subjects composed of virulent phages, the virome of IBD subjects is altered, less stable, and dominated by temperate phages. They show that changes in the virome reflect alterations in the bacteriome and that the use of both the bacteriome and virome composition was more effective at differentiating between IBD and healthy subjects (Figure 1). The results were validated on a longitudinal ulcerative colitis cohort. This database-independent approach could be used to shed light on viral dark matter from many published studies.
What are the consequences in practice
These findings confirm that the human gut virome is altered in IBD and that this may be associated with members of the bacterial microbiota; this could be used as a diagnostic or even prognostic biomarker. In addition, the observed alterations suggest that the virome could be partly responsible for the alterations in the bacterial microbiota seen in IBD. To this end, the gut virome could therefore be a future target or a future therapeutic tool in IBD.
Conclusion
Using a database-independent method, the whole gut virome can be studied whereas usually only a minority is analyzed. While the core virome in healthy subjects consists of virulent phages, the virome in IBD is altered, less stable, and dominated by temperate phages. Using both the virome and the bacterial microbiota composition makes it possible to differentiate IBD patients from healthy subjects more effectively than by using only one or the other. These results open new perspectives for the use of the virome as biomarker or therapeutic target in IBD.