Parkinson’s disease: discovery and inhibition of levodopa metabolism by gut bacteria
Commented articles - Adult's section
By Pr. Harry Sokol
Gastroenterology and Nutrition Department, Saint-Antoine Hospital, Paris, France
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections

Levodopa (L-DOPA) Parkinson's disease drug, molecular model. L-DOPA is a precursor of the catecholamine neurotransmitters dopamine, norepinephrine and epinephrine and is used in the treatment of Parkinson's disease. Atoms are represented as wires. A map of electrostatic potential surrounds the molecule.
About this article
Author
Commentary on the original publication by Rekdal et al. (Science 2019)
The human gut microbiota metabolizes the Parkinson’s disease medication Levodopa (L-dopa), potentially reducing drug availability and causing side effects. However, the organisms, genes, and enzymes responsible for this activity in patients and their susceptibility to inhibition by host-targeted drugs are unknown. Here, the authors describe an interspecies pathway for gut bacterial L-dopa metabolism. Conversion of L-dopa to dopamine by a pyridoxal phosphate-dependent tyrosine decarboxylase from Enterococcus faecalis is followed by transformation of dopamine to m-tyramine by a molybdenum-dependent dehydroxylase from Eggerthella lenta. These enzymes predict drug metabolism in complex human gut microbiotas. Although a drug that targets host aromatic amino acid decarboxylase does not prevent gut microbial L-dopa decarboxylation, the authors identified a compound that inhibits this activity in Parkinson’s patients microbiotas and increases L-dopa bioavailability in mice[1].
What do we already know about this subject?
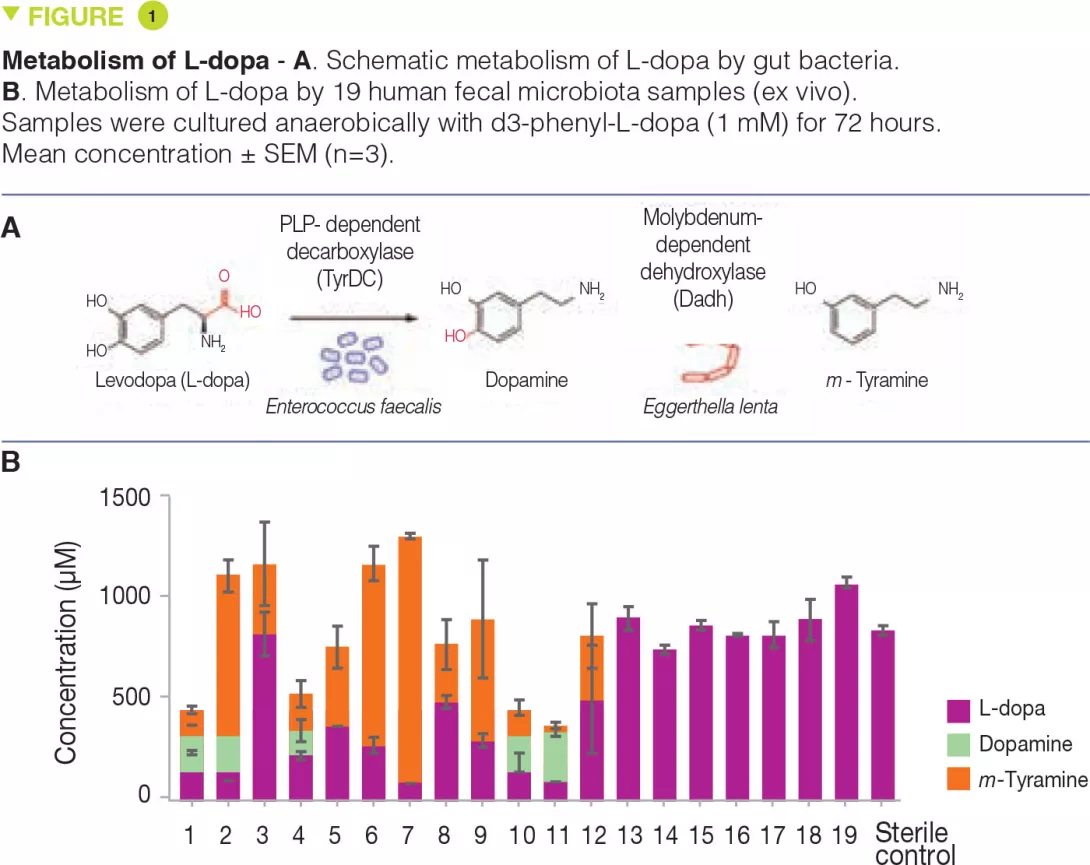
Parkinson’s disease is a debilitating neurological condition affecting more than 1% of the global population aged 60 and above. The primary medication used to treat Parkinson’s disease is levodopa (L-dopa). [2] To be effective, L-dopa must enter the brain and be converted to the neurotransmitter dopamine by the human enzyme aromatic amino acid decarboxylase (AADC). However, the gastrointestinal tract is also a major site for L-dopa decarboxylation, and this metabolism is problematic because dopamine generated in the periphery cannot cross the blood-brain barrier and causes unwanted side effects. Thus, L-dopa is co-administered with drugs that block peripheral metabolism, including the AADC inhibitor carbidopa. Even with these drugs, up to 56% of L-dopa fails to reach the brain. Moreover, the efficacy and side effects of L-dopa treatment are extremely heterogeneous across Parkinson’s patients, and this variability cannot be completely explained by differences in host metabolism. Previous studies in humans and animal models have demonstrated that the gut microbiota can metabolize L-dopa.[3] The major proposed pathway involves an initial decarboxylation of L-dopa to dopamine, followed by conversion of dopamine to m-tyramine by a dehydroxylation reaction.
Although these metabolic activities were shown to occur in complex gut microbiota samples, the specific organisms, genes, and enzymes responsible were unknown. The effects of host-targeted inhibitors such as carbidopa on gut microbial L-dopa metabolism were also unclear. As a first step toward understanding the gut microbiota’s effect on Parkinson’s disease therapy, the authors sought to elucidate the molecular basis for gut microbial L-dopa and dopamine metabolism.
Key points
-
Some gut bacteria can metabolize L-dopa to dopamine and then to m-tyramine, limiting its availability in the brain.
-
The gut microbiota plays a role in the efficacy and toxicity of L-dopa treatment for Parkinson’s disease.
-
Use of a specific inhibitor of bacterial L-dopa metabolism can increase L-dopa bioavailability and thus enhance its efficacy
What are the main insights from this study?
The authors hypothesized that L-dopa decarboxylation would require a pyridoxal phosphate (PLP)-dependent enzyme. They searched gut bacterial genomes and identified a conserved tyrosine decarboxylase (TyrDC) in Enterococcus faecalis (Figure 1A). Genetic and biochemical experiments revealed that TyrDC simultaneously decarboxylates both L-dopa and its preferred substrate, tyrosine. Next, they used enrichment culturing to isolate a dopamine- dehydroxylating strain of Eggerthella lenta (Figure 1A). Transcriptomics analysis showed that the enzyme responsible for this activity is a molybdenum cofactor-dependent dopamine dehydroxylase (Dadh). Unexpectedly, the presence of this enzyme in gut bacterial genomes did not correlate with dopamine metabolism. Instead, it is a single-nucleotide polymorphism (SNP) in the dadh gene that predicts activity. L-dopa metabolism through this pathway was variable across subjects (Figure 1B). In gut microbiotas from Parkinson’s patients, the abundance of E. faecalis, TyrDC, and the SNPs of dadh correlated with L-dopa and dopamine metabolism, confirming their relevance. The authors then showed that the human AADC inhibitor carbidopa had only a minimal effect on L-dopa decarboxylation by E. faecalis, and was completely ineffective in complex gut microbiotas from patients, suggesting that this drug likely does not prevent microbial L-dopa metabolism in vivo. Given TyrDC’s preference for tyrosine, the authors examined tyrosine “mimics” and identified (S)-α-fluoromethyltyrosine (AFMT) as a selective inhibitor of gut bacterial L-dopa decarboxylation. Co-administering AFMT with L-dopa and carbidopa to mice colonized with E. faecalis increased the serum concentration of L-dopa.
What are the consequences in practice?
These findings show that the gut microbiota can metabolize L-dopa and thereby influence the effectiveness and side effects of this drug. This study paves the way towards the discovery of predictive biomarkers for L-dopa efficacy and toxicity. Furthermore, since the underlying molecular mechanisms are known, the use of specific inhibitors of gut microbial L-dopa metabolism may be possible in patients whose microbiota contains bacteria with deleterious activities.
Conclusion
The gut microbiota in some Parkinson’s patients can metabolize L-dopa. This may underlie the heterogeneous efficacy and side effects of this treatment in Parkinson’s disease. The use of inhibitors of this bacterial metabolism could offer a solution. More generally, this study provides new evidence for the role of the gut microbiota in drug pharmacokinetics and pharmacodynamics. It opens up promising prospects for a field that could be called “pharmacomicrobiomics”.