Gut Microbiota #20
By Pr. Satu Pekkala
Academy of Finland Research Fellow, Faculty of Sport and Health Sciences, University of Jyväskylä, Finland
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections

About this article
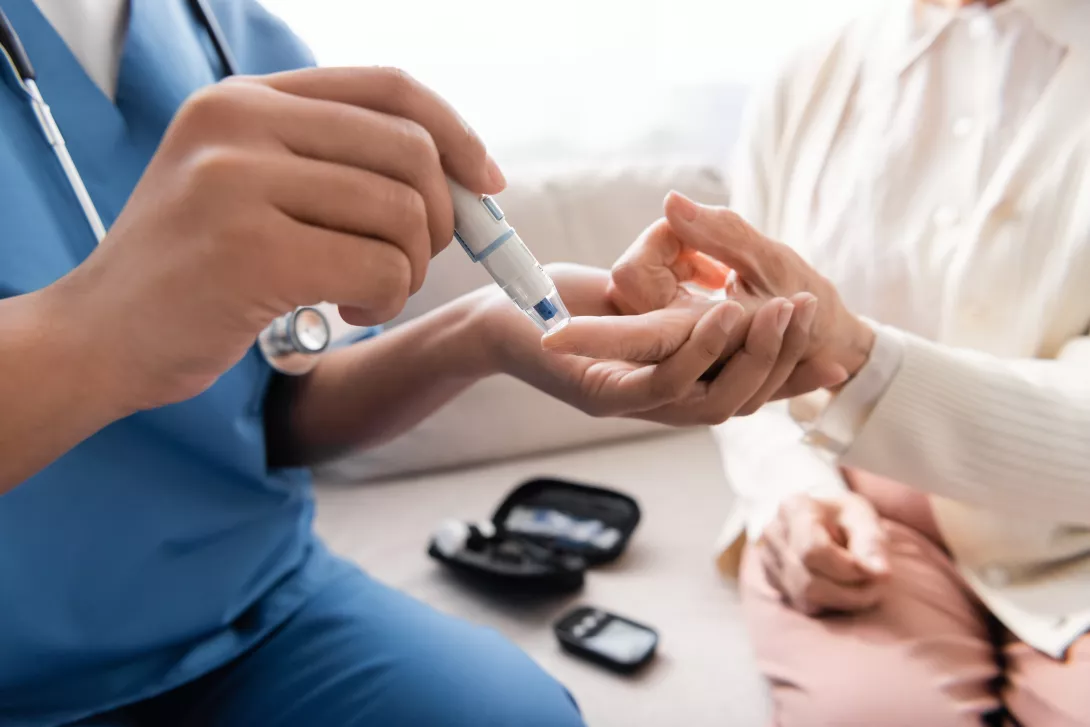
Microbiome-based personalized diet to improve pre-diabetes
Researchers at Weizmann institute have been pioneers in developing personalized dietary approaches based on gut microbiome (GM). In this paper, Ben-Yacov et al. studied the effects of personalized postprandial-targeting (PPT) versus Mediterranean diet (MED) on cardiometabolic risk factors. Overall, diet is known to affect cardiometabolic health but whether the GM modulates these effects has been scarcely studied in longitudinal settings. In this 6-months trial, 225 pre-diabetic adults were randomly assigned to PPT and MED arms. PPT was based on an algorithm and MED on dietitian judgement. Overall, there was a low-carbohydrate and high-fat pattern of the PPT intervention since dietary carbohydrate is an important component in post-prandial glucose response. Compared to MED, the PPT intervention increased GM diversity and richness more. In the PPT arm, the consumption of some catechin-rich foods including dark chocolate and cashews increased. This was further associated with the enrichment of Flavonifractor plautii that is reported to participate in flavonoid catechin metabolism. According to a statistical model, changes in specific GM species partially mediated the effects of diet on clinical outcomes. For instance, the change in UBA11471 sp000434215 (from Bacteroidales order) partially mediated the effect of change in ‘Med Oil and Fats’ consumption on HbA1c outcome. HbA1c is a glycated haemoglobin that is used to assess diabetes. Three bacterial species (from Bacteroidales, Lachnospiraceae and Oscillospirales orders) were found to mediate the effect of the PPT with clinical outcomes of HbA1c, HDL-cholesterol and triglycerides.
In conclusion, the study supports the role of GM in modifying the effects of diet changes on cardiometabolic outcomes and advance the concept of precision nutrition for reducing comorbidities in pre-diabetes.
Fecal metabolome in inflammatory bowel disease
Ulcerative colitis (UC) and Crohn’s disease (CD) are the subtypes of inflammatory bowel disease (IBD). Several microbial metabolites are known to affect inflammatory reactions that are important players in IBD. However, untargeted fecal metabolomic studies in IBD patients are scarce. In this study, the potential of fecal metabolites as biomarkers for IBD were assessed. The study comprised of 255 healthy controls and 424 IBD patients. The untargeted metabolomic analyses were accompanied with gut microbiota composition, exome sequencing, and genomic array data analyses in both cohorts. The metabolomes of the IBD groups were characterized by depletion of vitamins and fatty acid-related molecules. In addition, IBD patients had higher levels of the phenolic compound p-cresol sulphate, which originates when gut bacteria ferment proteins. The patients with UC had lowest levels of fecal anti-inflammatory short chain fatty acids. To identify potential biomarkers, a machine learning approach was used to predict disease phenotypes. The ratio of sphingolipid and L-urobilin discriminated between IBD and non-IBD samples. In the patients with IBD, the increase of pathobionts co-occured with increased levels of sphingolipids, ethanolamine and primary bile acids. In CD patients, resection of the ileocecal valve was associated with changes in the levels of 212 metabolites, such as cholic acid. Further, the resection associated in a reduced abundance of Faecalibacterium prausnitzii, which also negatively affected the levels of anti-inflammatory metabolites.
A mediation analysis showed that the observed associations between lifestyle, clinical factors and fecal metabolites were driven by alterations in the gut microbiota. Altogether, the study shows the potential of fecal metabolites as biomarkers for IBD and that, despite the influence of lifestyle, genetics and disease, gut microbes are strong predictors of the levels of fecal metabolites.
A microbiotamodulated checkpoint directs immunosuppressive intestinal T cells into cancers
Resistance of cancers to immune checkpoint inhibitors (ICIs) as a treatment can result from antibiotic (ABX) treatment, which may involve the gut microbiota. However, this relationship has not been extensively studied. Therefore, Fidelle and co-workers addressed the gaps in the knowledge using a rodent model and human patients. Based on the literature, the gut bacteria can induce the differentiation of lymphocytes primed in the mesenteric lymph nodes or homing to the intestinal lamina propria express the α4β7 integrin interacting with its counter-receptor, mucosal addressin cell adhesion molecule-1 (MAdCAM-1), which is expressed in high endothelial venules (HEVs). This importantly prevents the immigration of Treg17 cells to the gut tumors, which can further compromise the anticancer effects of ICIs. Th17 are a subset of pro-inflammatory T helper cells defined by their production of interleukin 17 (IL-17). These cells are related to T regulatory cells and the signals that cause Th17s to inhibit Treg differentiation. In rodents, ABX-treatment reduced MAdCAM-1 expression. This could be explained by the intestinal recolonization by the genus Enterocloster. Further, the oral administration of Enterocloster was sufficient to down-regulate MAdCAM-1 expression. Restoration of MAdCAM-1 expression on the ileal HEV by fecal microbial transplantation or blockade of IL-17A reversed the inhibitory effects of ABX. Ectopic expression of MAdCAM-1 in the liver caused a local maintenance of enterotropic α4β7+ Treg17 cells.
This maintenance further reduced their accumulation in the tumors as well as improved the efficacy of the immunotherapy in mice. In the cohorts of lung, kidney, and bladder cancer patients, low serum levels of MAdCAM-1 had a negative prognostic impact. In conclusion, the MAdCAM-1– α4β7 axis should be preferably considered in cancer immunosurveillance.


