Helicobacter pylori and gastrointestinal microbiota
By Pr. Juozas Kupcinskas
Department of Gastroenterology and Institute for Digestive Research, Lithuanian University of Health Sciences, Kaunas, Lithuania
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections
About this article
Helicobacter pylori (H. pylori) affects around 50% of the global population and is the most common bacterial infection worldwide. According to the World Health Organization (WHO), H. pylori is classified as a Group 1 carcinogen, which can cause precancerous and cancerous stomach conditions, including gastric adenocarcinoma. It was previously believed that H. pylori was the sole microorganism living in the human stomach, but recent advances in research technologies have provided a better understanding of the gastric microbiome. H. pylori plays a pivotal role in shaping the microbial environment of the stomach. However, the “non-Helicobacter” microbiome of the stomach has also been described and is a hot topic in current research. It is clear that H. pylori is a major modifier of the gastric microbiome, but other species may also contribute to carcinogenic pathways. The effects of proton-pump inhibitors (PPIs) and H. pylori eradication therapies on microbiome alterations have also been studied. In this review article, we aim to summarize recent findings on the gastric microbiome and the role of H. pylori in shaping it, as well as the impact of H. pylori eradication and PPIs on the human microbiome.
H. pylori is the major bacterium shaping gastric microbiome composition
The gastric microbiome is receiving increasing attention, with growing interest in its determining factors. Vilchez-Vargas et al. investigated microbial composition in several gastrointestinal (GI) compartments. Their study involved a cohort of 108 pairs of twins, and gastric microbiome biopsies from the stomach mucosae were analyzed. Microbiome diversity was assessed using V1-V2 regions of the 16S rRNA gene through amplification and sequencing. The results aligned with previous findings, highlighting H. pylori as a key factor in stomach microbiota composition 1.
Hua et al. conducted a study with a cohort of 193 patients to examine H. pylori’s impact on the richness and diversity of gastric microbiota in individuals with chronic gastritis. They profiled the V3- V4 region of the 16S rRNA gene and found significant alterations in gastric microbiota due to H. pylori infection. Specifically, H. pylori suppressed dominant gastric microbiota at the genus level, including Aliidiomarina, Reyranella, Halomonas, Pseudomonas, and Acidovorax. Their findings indicated that virulent strains of H. pylori were significantly associated with chronic atrophic gastritis and decreased gastric microbiota richness 2.
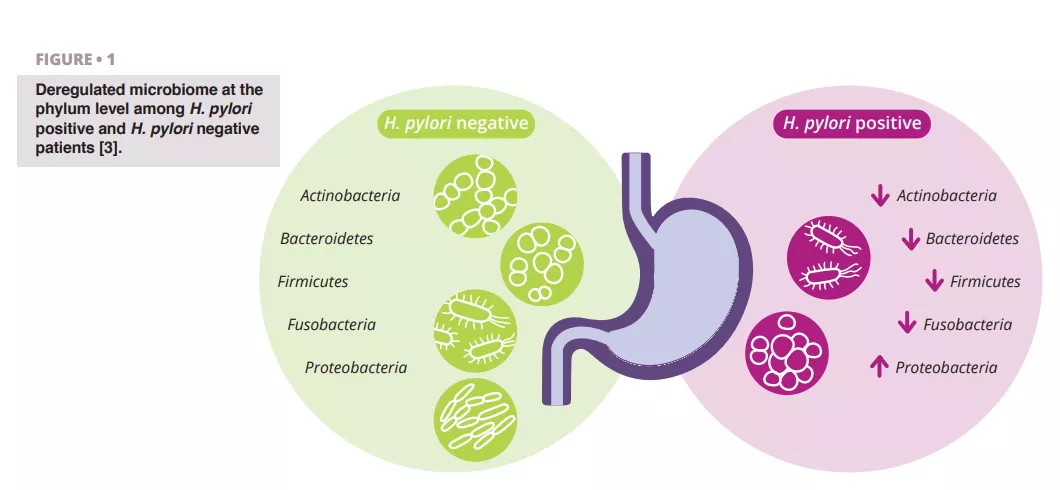
Schulz et al. analyzed differences in microbiota composition between H. pylori-infected and H. pylori-negative patients. They observed a significant difference in the relative abundance of Proteobacteria, which were more prevalent in the aspirates of H. pylori-infected patients. Other phyla showed lower relative abundance (figure 1) 3.
Miftahussurur et al. investigated gastric microbiota variability between H. pylori-positive and H. pylori-negative patients in a cohort of 137 individuals from the Indonesian population. They found that microbial β-diversity and richness were significantly higher in H. pylori-positive samples compared to H. pylori-negative samples. Additionally, their findings suggested that H. pylori plays a leading role in shaping the gastric microbial community in this ethnic group 4.
These studies collectively highlight H. pylori as a crucial factor in influencing gastric microbiome diversity and richness in the harsh stomach environment.
What is a true gastric microbiome?
The stomach presents extreme conditions for living microorganisms. A few decades ago, H. pylori was identified as a bacterium that could withstand these harsh stomach conditions. This discovery sparked curiosity and led to further research into the gastric microbiome. It remains uncertain whether non-Helicobacter bacteria in the stomach represent transient contaminants or part of a persistent microbiota. Spiegelhauer et al. conducted a study involving 22 patients with dyspepsia and 12 with gastric adenocarcinoma 5. They took biopsies from the stomach mucosa and analyzed the V3-V4 region of the 16S rRNA gene, in addition to culturing microorganisms. The authors hypothesized that H. pylori is the only true resident bacterium of the stomach and would persist in washed biopsies. Their findings indicated that bacterial load decreased in washed biopsies, suggesting transient contamination from the oral cavity. However, the diversity of microorganisms did not differ between unwashed and washed biopsies.
It remains unclear whether non-H. pylori microorganisms in the stomach are transient contaminants or genuine residents.
It is possible that continuously swallowed saliva, containing living organisms, may survive acidic conditions for a time. Contamination from the upper oropharyngeal region during gastroscopy and sampling should also be considered 6. These findings highlight the need for further investigation into the true gastric microbiome.
Effect of H. pylori eradication on gastric and gut microbiome
H. pylori is one of the most widespread infections globally, affecting more than half of the human population. Most treatment regimens for H. pylori involve two or more antibiotics, which can impact the gastrointestinal microbiome. Liou et al. investigated long-term changes in gut microbiota following H. pylori eradication. Their multicenter, randomized trial included 1,620 participants who were randomly assigned to three treatment groups. The authors assessed bacterial diversity at various times after eradication by analyzing fecal samples. Results showed that both alpha and beta diversity decreased within two weeks after eradication but returned to baseline levels by week 8 and one year later. These findings indicate only a shortterm perturbation of the gastrointestinal microbiome and suggest that H. pylori eradication therapy is generally safe in the long term 7.
He et al. reported on alterations in the gastrointestinal microbiome following H. pylori eradication, using 16S rRNA gene analysis of gastric mucosal and fecal samples. They found that alpha diversity of the gastric microbiome increased, and the beta diversity of the gut microbiome significantly differed from pre-treatment levels but resembled that of healthy controls 24 weeks after eradication 8.
Guo et al. summarized findings on changes in the gastric microbiome after successful H. pylori treatment. Their systematic review and meta-analysis included nine studies with 546 patients. This meta-analysis is the first to detail changes in alpha diversity after H. pylori eradication. Results indicated no significant differences in microbiota diversity between treatment options, whether quadruple or triple therapy was used. The authors observed increased alpha diversity in the short term, which persisted in long-term follow-up, with a depletion of H. pylori-related taxa and enrichment of common gastric commensals 9. To evaluate the effect of H. pylori eradication on the gut microbiome, Yap et al. conducted a study with 17 young adults. They sequenced the V3-V4 region of the 16S rRNA gene and analyzed the gut microbiome before and 18 months after H. pylori eradication using clarithromycin and metronidazole. No significant changes in microbial diversity were observed between baseline and 18 months post-eradication 10.
PPIs are major modifiers of the gut microbiome
Global consumption of proton pump inhibitors (PPIs) is increasing, making them one of the top 10 most widely used drugs in the world 11. They are a first-line treatment for conditions such as gastroesophageal reflux disease, peptic ulcer disease, dyspepsia, and, when combined with antibiotics, for the treatment of H. pylori 12. PPIs are often used without evidence-based indications or for longer periods than prescribed, and their use has been associated with an increased risk of infections such as Clostridium difficile, Salmonella spp., Shigella spp., Campylobacter spp., and other enteric pathogens 11.
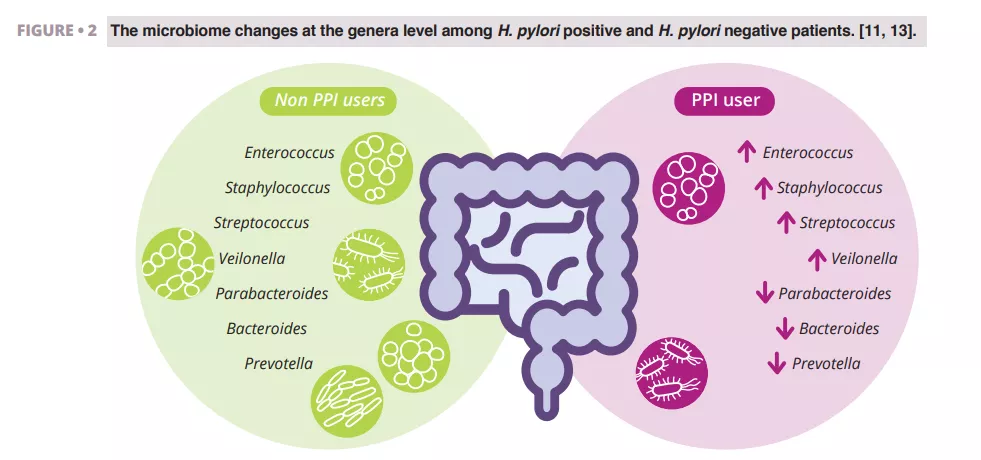
Imhann et al. analyzed the microbiome of 211 subjects using PPIs by sequencing the V4 region of the 16S rRNA gene. They observed a significant decrease in alpha diversity among PPI users and an increase in the abundance of bacteria from the genera Enterococcus, Streptococcus, Staphylococcus, and Veillonella (figure 2). The genera Enterococcus and Veillonella have been linked to a higher susceptibility to Clostridium difficile infections 11. While PPIs are generally considered safe with relatively rare side effects, evidence suggests they negatively impact the gut microbiome.
Zhang et al. conducted a meta-analysis of the effects of PPIs on human gut microbiota, analyzing data from four studies with 16S rRNA gene amplicon sequencing. Their results demonstrated a significant impact of PPI use on microbial diversity, with lower alpha diversity observed in PPI users compared to controls. They found decreases in the genera Parabacteroides, Veillonella, Bacteroides, and Prevotella, as well as from the families Ruminococcaceae and Lachnospiraceae (figure 2) 13.
Weitsman et al. performed a study with 177 subjects, matching PPI users 1:2 with controls. They analyzed stool samples and, for the first time, duodenal microbiomes. No significant differences in alpha or beta diversity were found between PPI users and controls. However, at the family level, they observed a higher relative abundance of Campylobacteraceae (phylum Proteobacteria) and a lower relative abundance of Clostridiaceae (phylum Firmicutes) in PPI users. Stool analysis similarly revealed a reduction in Clostridiaceae and an increase in Streptococcaceae 14.
Overall, these studies indicate that PPIs affect the human microbiome. The clinical significance of these findings warrants further investigation in future studies.

Conclusion
The human stomach presents a harsh environment for living microorganisms, and recent research continues to explore whether the stomach microbiota is persistent or merely reflects transient microorganisms. Despite its clinical significance, H. pylori plays a notable role in shaping the stomach’s microbiome. The eradication of H. pylori appears to have only a temporary and reversible impact on the composition of the gastric and gut microbiomes. Proton pump inhibitors (PPIs) are among the most commonly consumed medications. Scientific studies indicate that PPIs alter the structure of the gut microbiome; however, the clinical significance of these changes requires further investigation.

