Esophageal microbiome cause or consequence esophageal diseases
Press review
By Pr. Markku Voutilainen
Turku University Faculty of Medicine; Turku University Hospital, Department of Gastroenterology, Turku, Finland
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections
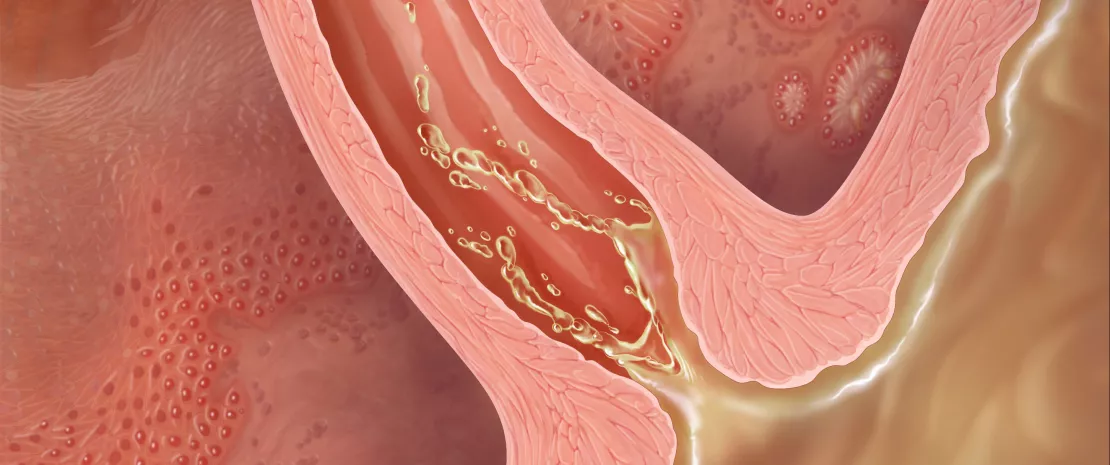
Illustration showing gastroesophageal reflux.
About this article
Author
Gastroesophageal reflux disease is common in western world. Barrett’s esophagus (BE) is the complication of reflux disease and the major risk factor of esophageal adenocarcinoma, which has a five-year survival rate less than 20%.
Recent review examined the role of esophageal microbiome in BE and esophageal cancer.[1] Esophagus is exposed to swallowed oral microorganisms and also microbes of the refluxed gastric contents. Esophageal microbiota is not similar to oral nor gastric microbiota. The first bacteria detected in the esophagus were Strectococcus viridans and group D Streptococcus. Later six phyla were observed by broad-range 16S ribosomal DNA gene clone sequencing including Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Streptococcus. BE and high-grade dysplasia were associated with the highest number of bacteria. In patients with esophagitis and BE the number of Streptococcus was diminished while gram-negative anaerobes and microaerophiles were increased.
BE and esophageal adenocarcinoma are associated with increased number of Escherichia coli. Another gram-negative species detected in esophageal cancer patients is Fusobacterium nucleatum. Also oral dysbiosis may be related to the increased risk of esophageal cancer, while gastric Helicobacter pylori seems to protect against esophageal cancer. Gastric dysbiosis such as increase of Clostridiales and Erysipelotrichaceae is associated with esophageal squamous carcinoma. Also fungi, for example Candica albicans and C. glabrata are often detected in esophageal samples from patients with esophaghageal adenocarcinoma. An epidemiologic study showed dose-dependent association between penicillin use and increased risk of esophageal cancer. Also proton pump inhibitors modify gastric and esophageal microbiome.
The present data of esophageal microbiota were obtained from small, selected, and symptomatic patient populations in cross-sectional studies. Thus no conclusions of causality between esophageal microbiota and esophageal diseases can be made. Only a small portion of patients with BE develop adenocarcinoma and further studies are needed to define the role of esophageal dysbiosis in the pathogenesis of cancer. One topic for further research is the impact of proton pump inhibitors on esophageal microbiota and on the risk for esophageal diseases.[1]
Eosinophilic esophagitis (EoE) is an allergic chronic inflammatory disease that is the most common cause of dysphagia in children and young adults in developed countries. EoE has common inflammatory features with other allergic diseases and allergen exposure probably has a central role in EoE pathogenesis. Capucilli and Hill have reviewed EoE epidemiology, pathogenesis and treatment.[2] The esophageal microbiota may be involved in the pathogenesis of EoE. The esophagus is colonized by hundreds of bacterial species and members of Firmicutes and Bacteroidetes phyla are the commonest [2]. In patients with active EoE, the genera Streptococcus and Atopobium are decreased while Neisseria and Corynebacterium are increased. Another study showed that the total amount of esophageal bacteria and Haemophilus genus specifically were increased in EoE. Proton pump inhibitors that are used in the treatment of EoE, cause an enrichment of Proteobacteria phylum. Esophageal bacterial load is increased in EoE patients irrespective of treatment or the severity of esophageal mucosal eosinophilia. Like in other allergic and autoimmune diseases, antibiotic treatment and caesarean delivery are associated with increased risk of EoE.[2]
Conclusion
Studies were cross-sectional and there is no data on the stability of esophageal microbiota over time. More studies are needed to define the role of esophageal microbiota in the pathogenesis and activation of EoE.